
Aditya Dendukuri
PhD Candidate
Department of Computer Science
University of California, Santa Barbara
E-mail: aditya_dendukuri [at] ucsb [dot] edu
About Me & Research Interests
My research lies at the intersection of applied mathematics, scientific computing, and data science, with a focus on stochastic modeling of discrete systems, rare-event sampling, and scalable algorithms for high-dimensional problems. I am particularly motivated by interdisciplinary collaboration and enjoy combining rigorous mathematical analysis with modern computational methods to tackle challenging scientific questions.
Current Projects
1. Efficient Stochastic Simulation Methods for Discrete Systems
I develop algorithms for simulating stochastic biochemical networks governed by the Chemical Master Equation:
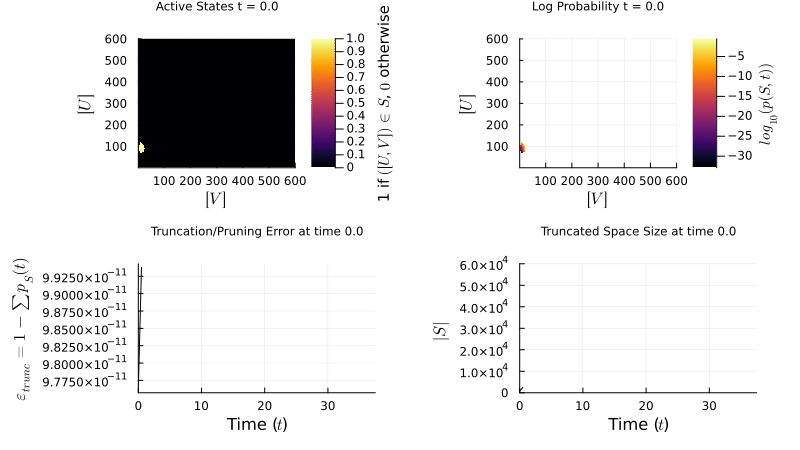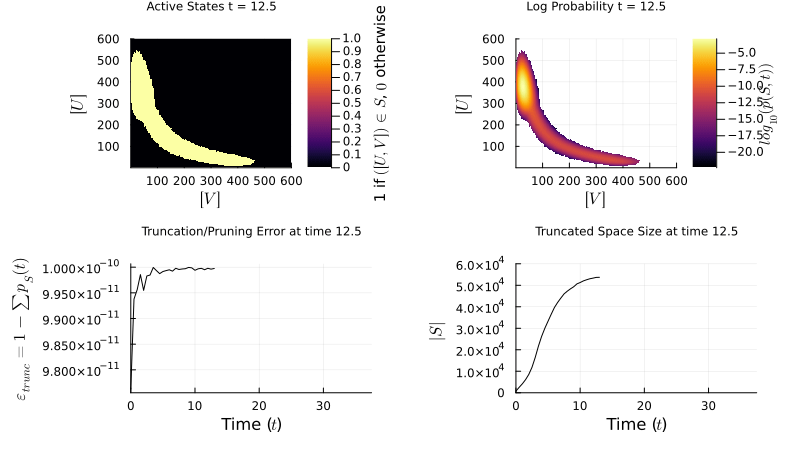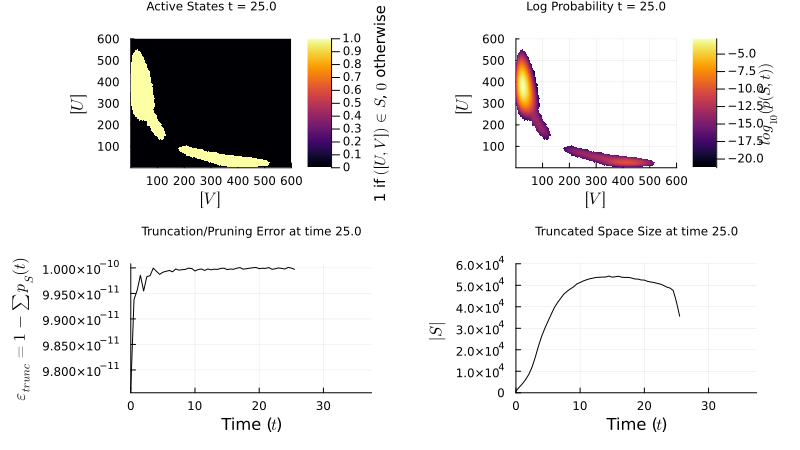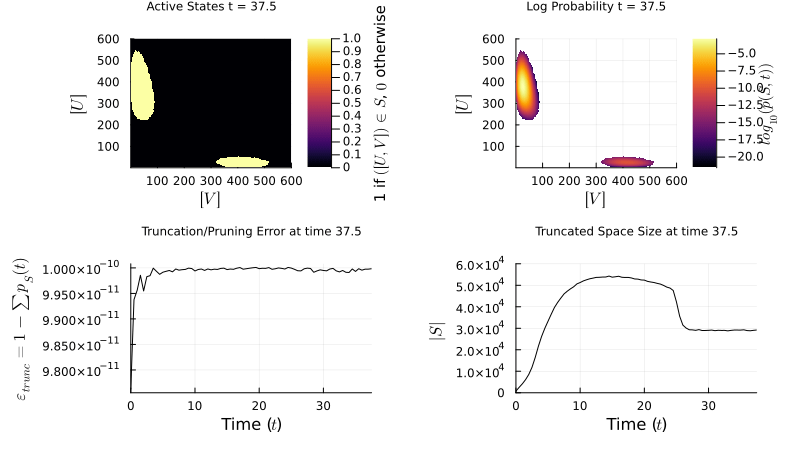 I implement the Finite State Projection (FSP) method to manage large state spaces:
I implement the Finite State Projection (FSP) method to manage large state spaces:
I developed Monte Carlo simulations of the Ising model to study nucleation—the initial stage of phase transitions such as crystallization. This C-based implementation applies umbrella sampling to construct free energy landscapes and capture rare nucleation events. The simulation is validated against theoretical predictions and includes visualization through ffmpeg integration. GitHub Repository